






Study with the several resources on Docsity

Earn points by helping other students or get them with a premium plan


Prepare for your exams
Study with the several resources on Docsity

Earn points to download
Earn points by helping other students or get them with a premium plan
Community
Ask the community for help and clear up your study doubts
Discover the best universities in your country according to Docsity users
Free resources
Download our free guides on studying techniques, anxiety management strategies, and thesis advice from Docsity tutors
It is Final Lab Report which is proposed by Afua Williams
Typology: Lab Reports
1 / 9

This page cannot be seen from the preview
Don't miss anything!






Bio 3302 - Final Lab Report 5/14/ Introduction: The identification of bacteria is important in order for us to differentiate one microorganism from another and classify them based on their morphology, ecology, genetics, and most importantly by species. Knowing bacterial species is especially useful because we can determine how different bacteria affect our existence, as some bacteria may be disease causing while others may be beneficial to the wellbeing of humans and our ecosystems. As a result, the identification of bacteria is very significant in the medical and pharmaceutical industries. The purpose of this experiment was to use techniques learnt throughout the semester to successfully identify two unknown bacteria, one gram-positive and one gram-negative, mixed in a broth culture in test tube 3. Materials and Methods: The methods below were followed from those listed in the course laboratory manual. An unknown bacterial culture in a test tube labeled 3 was chosen. Using aseptic techniques, streak plates were made and incubated for two days each on the following plates:
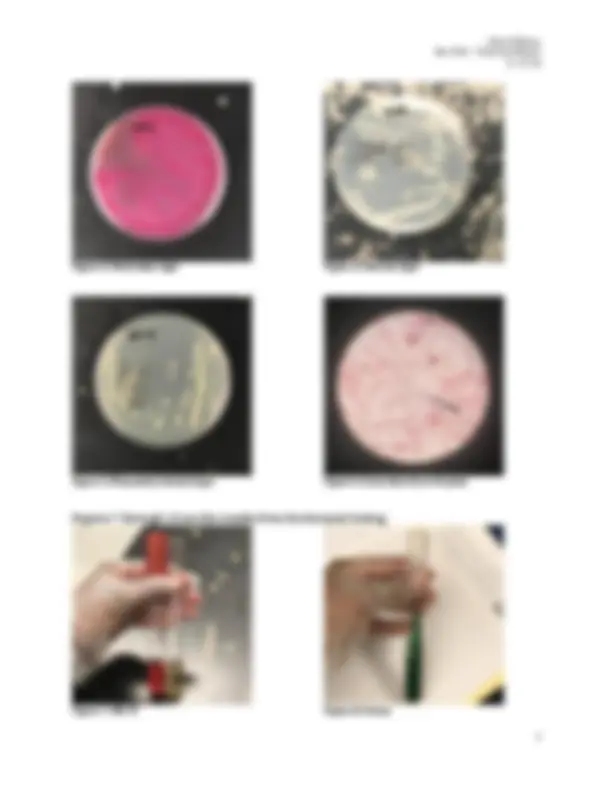
Bio 3302 - Final Lab Report 5/14/ Figure 3 : MacConkey Agar Figure 5 : Phenylethyl Alcohol Agar Figure 4 : Nutrient Agar Figure 6: Gram Stain from NA plate Figures 7 through 10 are the results from biochemical testing. Figure 7 : MR-VP Figure 8 : Citrate
Bio 3302 - Final Lab Report 5/14/ Figure 9: TSIA Figure 10 : SIMS The charts below were prepared and used as references throughout the experiments: Chart 1 : Gram Negative Bacteria and Their Characteristics
Bio 3302 - Final Lab Report 5/14/ Staphylococcus because there were colonies present in the MS plate. MS is also a differential medium used to identify mannitol fermenters that will produce acid and turn the medium yellow. Based on this knowledge, I was able to determine that the gram-positive bacterium was Staphylococcus aureus because of the two Staphylococcus species studied, it is the mannitol fermenter and the MS plate was almost completely yellow after the incubation period as seen in Figure 1 of the results. No further testing was necessary for identification. The unknown gram-negative bacterium was identified as Escherichia coli. MacConkey Agar was first used to isolate the gram-negative bacterium because it inhibits the growth of gram-positive bacteria. MAC is also differential in identifying gram-negative lactose fermenters that produce a pink color. Following the incubation period the results as seen in Figure 3 determined the bacterium was a lactose fermenter. Through this observation and based on the gram-negative bacteria studied, I was able to narrow down my gram-negative bacterium to Enterobacter aerogenes , Escherichia coli , or Serratia marcescens. After observing my nutrient agar plate (Figure 4 ), I did not think the bacterium was Serratia marcescens because it would have grown as reddish-brown colonies. The blood agar plate (Figure) also led me to believe that the bacterium was E. coli because of the beta hemolytic action observed. However, the MR- VP, TSIA, SIMS, and citrate biochemical tests were used for further identification because I was still uncertain. The ACID/ACID results from the TSIA (Figure 9) allowed me to safely eliminate S. marcescens because it is ALKALINE/ACID. The remaining test results were all characteristic of E. coli with the exception of the indole test which was negative when performed in the SIMS tube. This may have been due to cross contamination upon inoculation. The negative citrate test (Figure 8), and positive MR/ negative VP tests (Figure 7) results were however used to successfully identify the gram-negative bacterium as Escherichia coli.
Bio 3302 - Final Lab Report 5/14/ Staphylococcus aureus, according to the Centers for Disease Control (CDC), is a bacterium that resides in the noses of approximately 30% of the human population. It also can be found in the skin and gastrointestinal tract. Majority of the time S. aureus is not harmful but in healthcare settings however it may become infectious and even lead to death. It can travel into the bloodstream of individuals causing sepsis or bone infection. S. aureus is also known to be resistant to certain antibiotics which include Methicillin and Vancomycin. Individuals that are often at risk for staph are those with conditions like diabetes, eczema, and cancer. Escherichia coli are a diverse group of bacteria that are most often harmless. It is normally found living in the intestines of healthy individuals. Some strains of E. coli however can be infectious as a result of consuming contaminated food or water and can cause severe abdominal pain, excessive vomiting, and bloody diarrhea according to the CDC. According to Foodsafety.gov, the most toxic strain of E. coli is O157:H7, which produces the Shiga toxin and can cause kidney failure and death. E. coli was also the first bacterium used to study the stringent response in bacteria.