





Study with the several resources on Docsity

Earn points by helping other students or get them with a premium plan


Prepare for your exams
Study with the several resources on Docsity

Earn points to download
Earn points by helping other students or get them with a premium plan
Community
Ask the community for help and clear up your study doubts
Discover the best universities in your country according to Docsity users
Free resources
Download our free guides on studying techniques, anxiety management strategies, and thesis advice from Docsity tutors
Material Type: Lab; Professor: Moran; Class: Introductionto Biochemistry Laboratory; Subject: Chemistry; University: University of Wisconsin - Milwaukee; Term: Unknown 1989;
Typology: Lab Reports
1 / 8

This page cannot be seen from the preview
Don't miss anything!





A 280nm NaCl (mM) Fraction 40 KDa
30 KDa
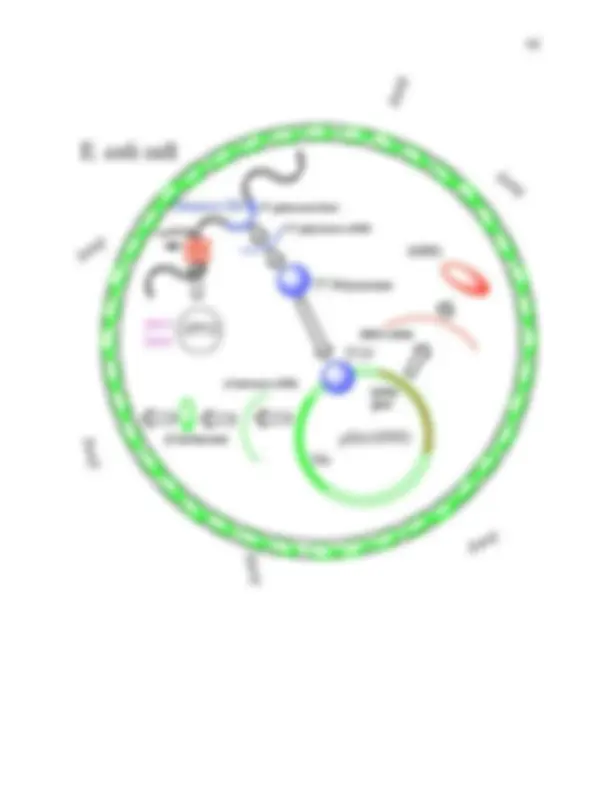
Experiment 2a INOCULATION OF LARGE-SCALE CULTURES In this short lab, each researcher will inoculate a fresh large-scale culture with the small- scale cultures grown in the previous lab. This large scale culture will then be used in the subsequent protein purification steps to isolate bacterial HPPD. Antibiotic resistance in bacteria is a serious problem facing society today. There are many reasons for this problem, one of which is an overuse of antibiotics. In hospitals where many people have compromised immune systems, resistant bacteria often kill a people with otherwise minor ailments. β-lactam antibiotics, such as the penicillins and the cephalosporins, are among the most commonly used antimicrobial agents. The production of β-lactamases, which catalyze β-lactam hydrolysis, is the predominant mechanism of bacterial resistance to these antibiotics. Here we select for those bacteria that can produce β-lactamase from a second gene in our recombinant plasmid and thus hydrolyse ampicillin (left) in the medium. The β-lactamase that is made is secreted into the periplasmic space and acts as a barrier against diffusing ampicillin (see Figure on the next page). In this way we allow only those cells that also contain the HPPD gene to grow since both genes are part of a single circular plasmid of DNA that we have constructed. Once the cells have reached a target density in the media, we will switch on the production of HPPD using a chemical known as iso-propyl-β-galactopyranoside (IPTG). The pET17b plasmid that we placed our HPPD gene in, is one of a series of plasmids that use the pET expression system. The pET System is the most powerful system yet developed for the cloning and expression of recombinant proteins in E. coli. Target genes are cloned in pET plasmids under control of strong bacteriophage T7 transcription. Expression is induced by providing a source of T7 RNA polymerase in the host cell. This is the function of IPTG, which induces the production of T7 polymerase from a man made “lac” based operon inserted into the host’s ( E. coli ) genome. T7 RNA polymerase is so selective and so highly active that almost all of the cell's resources are converted to target gene expression; the desired product can comprise more than 50% of the total cell protein a few hours after induction. S CH 3 CH 3 O HO H N O NH 2 O
Today’s Strategy Inoculate large scale cultures Induce cells with IPTG and allow to express HPPD Harvest cells by centrifugation Prepare Buffers for Experiments 2b and 2c
Record and Plot your Cell Growth Time (min) OD (600nm) Time (min) OD (600nm) -0. 0
1
2
3
-100 0 100 200 300 400 500 OD 600nm Time (min)
Prepare 1.5 L of 50 mM HEPES buffer.