



Study with the several resources on Docsity

Earn points by helping other students or get them with a premium plan


Prepare for your exams
Study with the several resources on Docsity

Earn points to download
Earn points by helping other students or get them with a premium plan
Community
Ask the community for help and clear up your study doubts
Discover the best universities in your country according to Docsity users
Free resources
Download our free guides on studying techniques, anxiety management strategies, and thesis advice from Docsity tutors
This document reports on a study conducted by Aimee Bakken et al. in which they injected cloned derivatives of Xenopus laevis ribosomal genes into X. laevis oocyte nuclei and examined the resulting transcription complexes in the electron microscope. The researchers concluded that the promoter lies between -320 and +113 nucleotides from the site of transcription initiation, and identified a fail-safe termination site just upstream from the initiation site.
Typology: Study notes
1 / 5

This page cannot be seen from the preview
Don't miss anything!




Proc. NatL Acad. Sci. USA Vol. 79, pp. 56-60, January 1982 Biochemistry
AIMEE BAKKEN, GARRY MORGAN, BARBARA SOLLNER-WEBBt,^ JUDITH^ ROANf,^ SHARON^ BUSBYt,^ AND RONALD H. REEDERt
*Department of Zoology,^ University^ of Washington, Seattle,^ Washington^ 98195;^ tDepartment of^ Physiological^ Chemistry,^ Johns^ Hopkins^ University Medical School, Baltimore, Maryland 21205; and tHutchinson Cancer Research Center, Seattle,^ Washington 98104 Communicated by Oscar L..Miller, Jr., September 1, 1981
the resulting transcription complexes in the electron microscope.
of the gene appears to^ be part^ of^ the normal^ termination^ signal.
nuclei and to assay transcription ofthese^ genes by spreading the
Gurdon (7), allows^ one^ to^ observe^ certain^ features^ of^ transcrip-
gene.
Barth's solution (11) at^ 0.5^ jug/gl, and^ in^ most^ experiments^ 5-
mixed with the DNA solution at (^200) Ag/ml. Injection of^ 10- nl of a-amanitin at this concentration was^ enough to^ greatly re- duce transcription from^ an^ injected plasmid carrying X.^ laevis 5S (^) genes (data not shown). Electron Microscopy. Nuclei of injected oocytes were man-
removed with forceps (6). The nuclear^ contents^ were^ spread for electron (^) microscopy essentially as described by, Miller and Bak-
scribing plasmid molecules. Preparations were^ stained^ in^ 1%
Cloning ofrDNA Fragments. All rDNA plasmids used^ in^ this
using P2-EK1 containment procedures required by the^ current
nuclei. pXlr 101A contains a full repeating unit of X. laevis
Abbreviations: kb, kilobase^ pairs; bp, base^ pairs;^ rDNA,^ ribosomal DNA. 56
The publication costs^ ofthis article^ were^ defrayed^ in^ part^ by^ page^ charge payment. This article^ must^ therefore^ be^ hereby^ marked^ "advertise- nent" in^ accordance^ with^18 U.^ S.^ C.^ §1734^ solely^ to^ indicate^ this fact.
Proc. NatL Acad. Sci. USA 79 (1982) 57
Inifiction i (^) BR
I~EcoRtI t , BomHIHindX t Psi S XorI
i
v--
pXivI4D
28S L (^) , pBR322 (^) ffi XrIA
.I pXlr2O 1 p)Ar Ikb
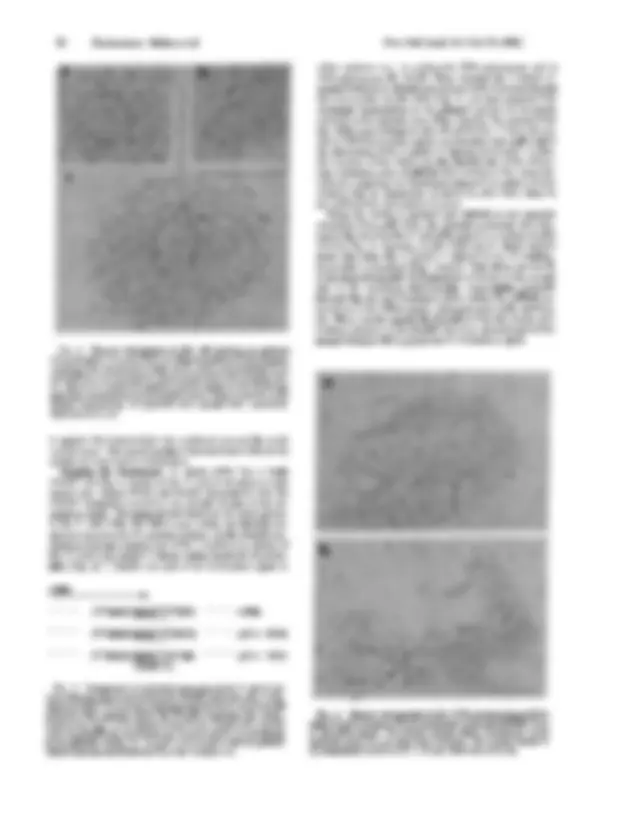
FIG. 1. Structure of various plasmids containing fragments of ri- bosomal genes. bp, Base pair; kb, kilobase pair. pXlr 14D: A 433-bp fragment bounded by a (^) Pst I site at -320 and an Xor II (^) site at + was removed from the parent pXlr 14 (13) and inserted between the Pst and Xor II sites in pBR322. pXlr lOlA: This plasmid contains a single full^ repeat unit of X.^ laevis rDNA inserted at the HindiI site of pBR322. The insert in this plasmid was originally cloned from a Hindl digest of amplified rDNA inserted into pMB9 and was termed pXlr 101. pXlr 101 is the plasmid that was used by Trendelenburg and Gurdon (7) for oocyte injection. We subsequently removed the insert from pMB9, put it in pBR322, and renamed it pXlr lOlA. The inserts in bothplasmids are in the same orientation relative to the tetracycline resistance gene. pXlr lOlB: This plasmid is identical to pXlr lOlA ex- cept that the rDNA insert is in the opposite orientation relative to the tetracycline resistance gene. pXlr 203: This plasmid contains a mini- gene that should produce a transcript about 2.75 kb in length. It was constructed by linking an EcoRI/Bamffl fragment from the 5' end (^) of the (^) gene to an EcoRI/BamHI (^) fragment from the 3' end of (^) the gene and inserting the result at the BamHI site of pBR322. The fragments were derived from pXlr 14 (13) and the initiation and termination sites are the ones whose sequences were determined by Sollner-Webb and Reeder (2). pXlr 203A150: This is a deletion mutant of pXlr 203 in which a segment from the BamHI site at -1150 before initiation to
scripts that are morphologically identical to the gradients seen on the^ endogenous ribosomal^ genes (Fig. 2). In^ good oocytes, we routinely see several hundred actively transcribing plasmids per electron^ microscope grid. Often the transcribed^ region is preceded by a nonbeaded spacer region, whereas the pBR portion of the circle^ remains^ beaded, in^ agreement with^ pre- vious observations (7). Table 1 tallies the RNA polymerase pack- ing density for both^ the^ injected and the^ endogenous ribosomal genes and shows that they are indistinguishable by this crite- rion. All (^) transcription units on (^) pXlr 101A molecules were of normal length and polymerase density.
with a-amanitin, the transcription complexes on the plasmids are completely normal-i.e., resistant to this inhibitor of RNA polymerases II and III. These data show that transcription on the injected rDNA is the result of specific initiation^ by RNA polymerase I^ and that this electron^ microscopy assay can^ be used to study the DNA sequences required for polymerase I promoter function. Transcription Initiation on pXlr.203 and pXlr 14D.^ To begin identification of the rDNA (^) sequences involved in promoter function we injected pXlr 203. This plasmid, which contains sequences from -1050 nucleotides before^ and +2250 nucleo- tides after the initiation site (see Fig. 1), reproducibly initiates transcription upon injection into oocytes (data not^ shown). In order to further map the^ promoter sequence we^ constructed pXlr 14D, a plasmid that contains a 433-bp rDNA insert ex- tending from a Pst I site at^ -320^ bp to^ an^ Xor^ II^ site^ at^ +^113 bp (see Fig. 1). As^ shown^ in^ Fig. 3, pXlr 14D^ is^ transcribed^ upon injection. The^ density of^ RNA^ polymerases on^ pXlr 14D^ was as high or^ higher than that^ seen on^ endogenous ribosomal^ genes
4e~~~~~~~~
r.0.
as~~~~~,thtse*nedoeosrbsmlee 24 .35 un=2)
(A~~ ~ ~ ~
(a) Example that clearly shows that the (^) nontranscribed spacer (to the right of^ the^ arrow)^ is smooth, while^ the pBR322^ vector (left of the^ ar row) is in a beaded configuration. (b) Example in which a-amanitin was injected with the DNA. Scale bar is 0.5 (^) gm.
(Table 1). Also, this^ transcription was^ not^ affected^ by a-aman-
the pBR322 vector.
Table 1. RNA polymerase density on endogenous ribosomal genes and recombinant plasmids RNA polymerase density, Source of rDNA (^) molecules/lpm Endogenous ribosomal genes 35. pXlr lOlA 34. pXlr l01B 35. pXlr 14D 42. At least 20 molecules of each type were measured.
't P-^ .- if^ v
i
Biochemistry: Bakken^ et^ al.
11 "I spocer)^ "I
;t
Proc. Natl. Acad. Sci. USA 79 (1982) 59
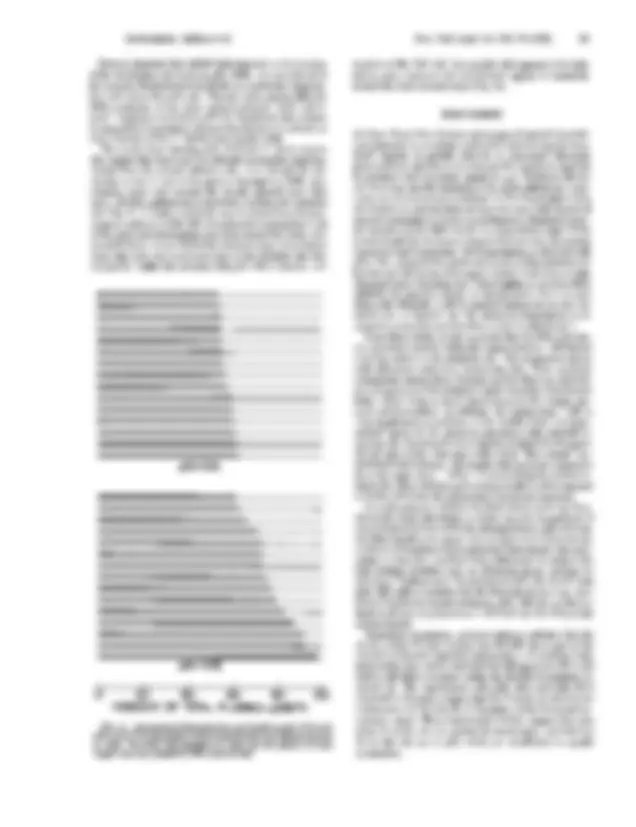
the nascent ribonucleoprotein fibrils on a particular transcrip- tion unit stop at the same site. This site varies among different DNA molecules of the same injected plasmid. Such "all-or- none" stopping is consistent with the hypothesis that a factor is required for termination (18) and that the factor is present on only a fraction of the 3' HindIII sites of pXlr 10iB. The results from injecting pXlr lOB into X. laevis oocytes also suggest that there may be a fail-safe termination signal up-
pXIrlOlA
-~~~~~~~~~~~~~~~~
pXlr l01lB #### PERCENT OF TOTAL PLASMID LENGTH FIG. 6. Comparison of transcription unit length on pXlrlOlA and pXlr (^) 10iB. The total length of each plasmid circle has been normalized to 100%. (^) The thick line represents in each case the percent of total length that was covered by RNA polymerases. ### mination at this "fail-safe" site on pXlr 14D appears to be leaky ### and in many instances the polymerases appear to transcribe ### around the circle several times (Fig. 3c). #### DISCUSSION ### We have shown that electron microscopy of injected recombi- ### nant plasmids is a workable method for determining the func- ### tional capacity of partially deleted or rearranged ribosomal ### genes and thus will allow us to map specific sequences required ### for promoter and terminator signals in vivo. Evidence that we ### are observing specific initiation at the RNA polymerase I pro- ### moter can be summarized as follows: (i) The transcription units ### we observe on injected plasmids have the same high density of ### nascent transcripts as is seen on endogenous ribosomal genes. (ii) Injection of pXlr 101A^ results in^ a transcribed region of the ### correct length for the known distance between the polymerase ### I promoter and termination. (iii) Transcription is observed with ### pXlr 14D, a plasmid that carries the in vivo rDNA initiation site ### but has only 433 bp out of the approximately 11 kb of an average ### ribosomal gene repeating unit. Transcription is not seen when ### pBR322, the plasmid vehicle, is injected alone. Nor is it seen ### when pXlr 203A150, a rDNA plasmid lacking the in vivo ini- ### tiation site, is injected. (iv) The observed transcription is re- ### sistant to a-amanitin and therefore is due to polymerase I. ### From these results we also conclude that the RNA polymer- ### ase I^ promoter must lie within the region between - 320 bp and ### + 113 bp relative to the initiation site. This assignment agrees ### with inferences made from sequencing data. First, sequence comparisons among three Xenopus species show no conserva- ### tion of sequences in the initiation region except for 15 bp located ### from -11 to +4 bp on the X. laevis sequence (M. Crippa, per- sonal communication). In addition, the region from -145 to ### +4 is duplicated several times in the middle of the "non-tran- scribed" spacer (2, 19), always in association with a BamHI re- striction site. Occasionally transcription is initiated in the spacer (20-22) and, in fact, does start within these "Bam islands" (un- ### published observations). This implies that promoter sequences lie in the region from -145 to +4 surrounding the normal ini- ### tiation site. Finer deletion and mutation studies will be required to further delineate the polymerase I promoter sequence. ### It is still unknown whether the Bam islands serve any func- ### tion in the frog's physiology or if they are only by-products of ### recombination events within the ribosomal locus. pXlr101A has two Bam islands in its spacer, but we have never observed any evidence of initiation at these particular Bam islands. One spec- ulation is that they somehow help polymerase to achieve the high packing densities seen on ribosomal genes, perhaps by ### serving as "loading zones" for polymerase (23). Our results with pXlr 14D make it unlikely that the Bam islands have any man- ### datory function in normal initiation. pXlr 14D has no Bam is- lands at all and yet polymerase I still loads on this DNA at the normal density. Regarding termination, present evidence (^) indicates that the cluster of four Ts that overlaps the HindIII site is part of the ### normal termination signal for polymeraseI. S1 nuclease map- ping studies have shown that both the 40S precursor (^) RNA and mature 28S RNA terminate within the HindIII (^) recognition se- quence (2). The experiments with pXlr 101A and pXlr (^) 101B ### reported in this paper suggest that the T cluster is not only the ### termination site but also the 3' boundary of the functional ter- ### mination signal. These experiments further suggest that only ### three Ts at this site are needed for termination, and that two ### Ts at this site (as in pXlr lOiB) are insufficient to specify termination. a m Biochemistry: Bakken^ et^ al. ### 60 Biochemistry: Bakken et aP ### It is important to note that termination of pXlr lOlA tran- ### scription is most likely occurring at the natural rDNA termin- ### tion site rather than at a fortuitous stop site in the adjacent ### pBR322 sequences. If such a fortuitous, high-efficiency ter- ### mination site in pBR322 formed the 3' boundary of the pXlr ### LOLA transcription unit, it would have to reside in the tetra- ### cycline resistance gene, downstream from the HindIII insertion ### site. Yet, this exact region is present in the same orientation ### relative to the rDNA in pXlr 14D-800 nucleotides downstream ### from the initiaion site (see Fig. 1)-and in this plasmid tran- ### scription always proceeds completely through the entire tet- ### racycline resistance gene without terminating (Fig. 3). ### In pXlr lOlA it appears that a cluster of three Ts at the ### HindIII site is sufficient for termination. However, it is very ### likely that some of the preceding nucleotides also are needed ### to specify termination in pXlr lOlA, because clusters of three ### Ts found at other, internal sites in the gene do not cause ter- ### mination (2, 4). ### The results with pXlr lOlB and pXlr 14D further suggest the ### presence ofa fail-safe termination signal(s) in front ofthe normal ### initiation site as noted above. The presence of a fail-safe ter- minator is supported also by (^) observations ofendogenous genes ### that occasionally show transcription complexes in the spacer ### region (20, 22). These "prelude complexes" always seem to ter- ### minate before running into the 5' end ofthe gene. Examination ### of the DNA sequences upstream from initiation shows two T ### clusters that are likely candidates for termination signals, one ### beginning at nucleotide -27^ and the other at -228^ (2). ### In this study, the intact rRNA gene found in pXlr LOlA [as ### well as in its parent plasmid, pXlr 101 (data not shown)] pro- duces only transcription units with a length identical to that of ### the endogenous ribosomal transcription units. In previous stud- ### ies in which pXlr 101 was injected into X. laevis oocytes (7) or ### into Triturus oocytes (24, 25), transcription units^ of other ### lengths were visualized also. At present, we can offer no ex- ### planation for these conflicting results. Regarding the frequency ### of transcription, we cannot make an accurate estimate of the ### fraction of injected plasmids that are transcribed. Even in the best (^) preparations both active and inactive (^) plasmids are often ### seen in large unresolvable aggregates. In addition, control ex- ### periments (unpublished) show that less than^ 10%^ ofthe^ injected ### DNA is deposited on the electron microscope grid. It^ is^ possible ### that plasmids densely packed with transcription complexes are ### preferentially deposited during centrifugation. However, be- ### cause we detect transcription only when a bona fide rDNA pro- ### moter is present in the plasmid, we are confident that the elec- ### tron microscope provides a reliable assay for promoter functions. ### We gratefully acknowledge the support of this research by National Institutes of Health Grants GM-28905 to A.B. and R.H.R. and GM- 27720 to B.S.-W. 1. Boseley, P., Moss, T., Machler, M., Postman, R. & Birnstiel, M. (1979) Cell 17, 19-31. 2. Sollner-Webb, B. & Reeder, R. H. (1979) Cell 18, 485-499. 3. Moss., T., Boseley, P. G. & Birnstiel, M. L. (1980) Nucleic Acids Res. 8, 467-485. 4. Course, R. & Gerbi, S. (1980) Nucleic Acids Res. 8, 3632-3637. 5. Reeder, R. H., Sollner-Webb, B. & Wahn, H. L. (1977) Proc. Natl Acad. Sci. USA 74, 5402-5406. 6. Miller, 0. L., Jr., & Bakken, A. H. (1972) Acta Endocrinot (Co- penhagen) Suppl 168, 155-177. 7. Trendelenburg, M. & Gurdon, J. (1978) Nature (London) 276, 292-294. 8. (^) Dumont, J. N. (^) (1972) J. Morphol. 136, 153-179. 9. La Marca, M.^ J., Smith, L. D.^ & Strobel, M. C. (1973) Dev. Biot 34, 106-118. ### 10. Hipskind, R. A. & Reeder, R.^ H. (1980) J. Biol Chem. 255, 7896-7906. 11. Gurdon, J. B. (1974) The Control of Gene Expression (Harvard Press, Cambridge, MA). 12. Kressman; A. & Birnstiel, M. L. (1980) in Transfer of Cell Con- stituents into (^) Eukaryotic Cells, eds. (^) Celis, J. E., Grassmann, A. & (^) Loyter, A. (^) (Plenum, New (^) York), Vol. (^) 31, pp. 383-407. 13. (^) Botchan, P., Reeder, R. H. & (^) Dawid, I. B. (^) (1977) Cell 11, 599-07. 14. Sutcliffe, J. G. (1978) Nucleic Acids Res. 5, 2721-2728. 15. Adhya, S. & Gottesman, M. (1978) Annu. Rev. Biochem. 47, 967-996. 16. Kom, L., Queen, C. & Wegman, M. (1977) Proc. Nati Acad. Sci. USA 74, 4401-4405. 17. Bogenhagen, D. F. & Brown, D. D. (1981) Cell 24, 261-270. 18. (^) Leer, J. C., Tiryaki, D. & (^) Westergaard, 0. (^) (1979) Proc. Natl. Acad. Sci. USA (^) 76, 5563-5566. 19. Moss, T. & Birnstiel, M. L. (1979) Nucleic Acids Res. 6, 3733-3743. 20. (^) Scheer, U., Trendelenburg, M., Krohne, G. & (^) Franke, W. (1977) Chromosoma 60, 147-167. 21. Rungger, D. & Crippa, M. (1977) Prog. Biophys. Molt Biol 31, 247-269. 22. Morgan, G., Bakken, A. & Reeder, R. (1981) (^) J. Cell Biol. 91, 365a. 23. Mueller, K., Oebbecke, C. & Forster, G. (1977) Cell 10, 121-130. 24. Sollner-Webb, B., Bakken, A., Morgan, G., Busby, S. & Reeder, R. (^) (1980) Carnegie Inst. (^) Washington Yearb. (^) 79, 70-72. 25. (^) Bakken, A., Morgan, G., Sollner-Webb, B., Roan, J., Busby, S. & (^) Reeder, R. (^) (1982) in (^) Perspectives in (^) Differentiation and (^) Hy- pertrophy, ed.^ Anderson, W.^ (Elsevier, New^ York), in^ press. Proc. Nad Acad. Sci. USA (^79) (1982)