





Study with the several resources on Docsity

Earn points by helping other students or get them with a premium plan


Prepare for your exams
Study with the several resources on Docsity

Earn points to download
Earn points by helping other students or get them with a premium plan
Community
Ask the community for help and clear up your study doubts
Discover the best universities in your country according to Docsity users
Free resources
Download our free guides on studying techniques, anxiety management strategies, and thesis advice from Docsity tutors
An in-depth analysis of the enzymes involved in DNA replication, including their functions, pictorial representation, and significance in DNA synthesis. the key enzymes such as DNA polymerase I, II, and III, as well as endonucleases, helicase, SSB protein, primase, and DNA ligase. It also discusses the role of these enzymes in DNA replication and repair, and their importance in maintaining the integrity of genetic material.
What you will learn
Typology: Slides
1 / 8

This page cannot be seen from the preview
Don't miss anything!





This document holds the enzymes used in DNA replication, their pictorial representation and functioning.
DNA polymerase is the chief enzyme of DNA replication. DNA polymerase activity was discovered by Arthur Kornberg in
All DNA polymerases require a template strand, which is copied. DNA polymerases also require a primer, which is complementary to the template. The reaction of DNA
polymerases is thus better understood as the addition of nucleotides to a primer to make a sequence complementary to a template. The requirement for template and primer are exactly what would be expected of a replication enzyme. Because DNA is the information store of the cell, any ability of DNA polymerases to make DNA sequences from nothing would lead to the degradation of the cell's information copy.
All DNA polymerases require the following:
(1) A template DNA strand, (2) A short primer (either RNA or DNA), and (3) A free 3′ -OH in the primer.
They add one nucleotide at a time to the free 3′ -OH of the primer, and extend the primer chain in 5′ → 3 ′ direction.
Figure 1 : Arthur Kornberg
single polypeptides, but DNA polymerase III is a ten subunits protein with a molecular mass of approx. 900KD.
DNA polymerase α catalyzes priming of both the strands. DNA polymerases ξ, η, τ, and k are all nuclear DNA repair enzymes. DNA polymerase γ is found in
mitochondria and catalyzes replication of mtDNA.
Enzyme # Endonucleases:
An endonuclease produces an internal cut (single- or double-stranded) in a DNA
molecule. But a restriction endonuclease produces cuts only at those sites that have a
specific base sequence. During DNA replication, an endonuclease may induce a nick to
initiate DNA replication, or it may induce nicks to generate a swivel for DNA unwinding.
Enzyme # DNA gyrase
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase (Type II
topoisomerase) that relieves strain while double-stranded DNA is being unwound by
helicase. This causes negative supercoiling of the DNA. The gyrase supercoils (or relaxes
positive supercoils) into DNA by looping the template so as to form a crossing, then
cutting one of the double helices and passing the other through it before releasing the
break, changing the linking number by two in each enzymatic step.
Enzyme # Helicase:
Helicase effects strand separation at the forks and use one ATP molecule for each base that is separated. In E. coli, DNA functions as helicase; this protein is a hexamer and it moves with the replication fork. Helicase breaks the hydrogen bonding between bases.
Enzyme # Single-Strand Binding (SSB) Protein:
SSB protein binds to single-stranded DNA, and prevents it from forming duplex DNA or
secondary structures. SSB binds as a monomer, but it binds cooperatively in that binding
of one SSB molecule facilitates binding of more SSB monomers to the same DNA strand.
E. coli SSB is a tetramer.
Enzyme # Polynucleotide Ligase:
DNA ligase or polynucleotide ligase catalyzes the formation of phosphodiester linkage between two immediate neighbour nucleotides of a DNA strand. Thus it seals the nicks remaining in a DNA strand either following DNA replication or DNA repair. However, this enzyme cannot fill the gaps in DNA strands.
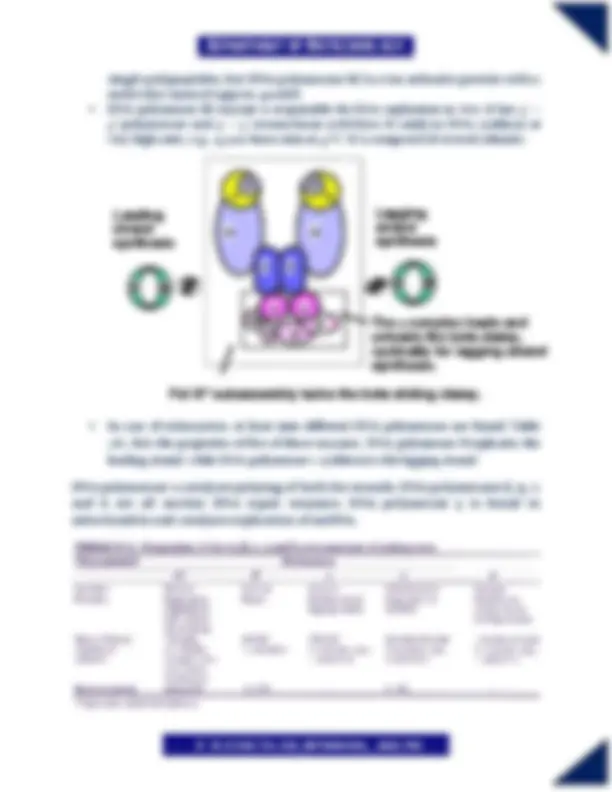
Figure: Stepwise action of enzymes involved in the replication
References